Abstract
Lipid nanoparticle-based messenger RNA (mRNA) delivery system have ushered in a new era of gene therapeutics. However, their clinical advances have been impeded by the inherent liver tropism of lipid nanoparticles (LNPs), which limits extrahepatic applications. Herein, we highlight two recent studies written in Nature Materials. Guided by surface interface molecular mechanics, Chang and his colleagues worked on the intelligent design of targeted peptides combined with lipids. Concurrently, Lin and his colleagues explored novel organ-specific peptide-lipid materials and analyzed important LNP traits that co-determined the in vitro/in vivo transfection discrepancy. The resulting peptide-modified lipids exhibit the capacity for targeted delivery to various organs and realize gene editing function. These studies jointly established a modular platform that provides a rational and predictable strategy for designing tissue-specific LNPs, thereby paving the way for next-generation mRNA-based therapeutics.
Keywords
1. Introduction
Lipid nanoparticles (LNPs) have become the primary delivery system for messenger RNA (mRNA) due to their high efficiency and low immunogenicity[1,2]. The success of COVID-19 vaccines has demonstrated their effectiveness, safety, and scalability for industrial production[3,4]. However, the broader therapeutic potential for LNP-mediated non-hepatic delivery is limited by predominant liver accumulation following systemic administration. Although strategies such as LNP composition optimization[5,6], lipid chemical modification[7,8], and ligand conjugation[9,10] have been explored to address this limitation, platforms that can cover diverse rationally designed lipids to enable the precise targeting of specific organs require further exploration.
Peptides offer great design flexibility and modular tunability based on the diverse physicochemical properties of their constituent amino acids[11]. Thus, peptides have been widely used in cell penetration[12], targeted delivery[13], and antitumor therapeutics[14]. Recently, through the integration of functional peptides into LNPs and modular tuning of amino acid sequences, both Chang’s[15] and Lin’s[16] teams have developed peptide-modified LNPs (PM-LNPs) that exhibit enhanced tissue-specific targeting and strengthened mRNA delivery efficiency. They approached this goal from distinct angles: Chang’s work employed rational design, elucidating the targeting mechanism to build an artificial intelligence (AI)-assisted framework, while Lin’s research developed the solid-phase support synthesis (SPSS) strategy for customizable synthesis and tried to understand the connection between different performances of LNPs in vitro and in vivo. Their complementary contributions offer a promising strategy to achieve improved organ-selective mRNA delivery.
2. Universal Construction of PM-LNPs
PM-LNPs consist of three core components: peptide-modified lipids (PMLs), structural lipids, and nucleic acid molecules. PMLs are formed by covalent conjugation between peptide heads and lipid tails. Lin’s approach involves SPSS, which directly links natural individual amino acids or other functional blocks to artificial ionizable alkylated Fmoc-protected amino acids (AIFAs)[16]. At the same time, Chang’s work chose to conjugate short peptides to PEGylated lipids via click chemistry to achieve the same goal[15]. PMLs, together with structural lipids, self-assemble in a microfluidic chaotic mixer. The resulting LNPs are capable of efficiently encapsulating and delivering various nucleic acid payloads, including mRNA, engineered prime editing guide RNA (pegRNA), and single-guide RNA (sgRNA), enabling organ-specific targeted gene editing.
Rapid construction of a diverse PML library serves as a critical foundation for accelerating the screening of latent lipids for clinical use. Computer-aided design, widely employed in protein generation, evaluation, and optimization[17], can be leveraged to rationally design and screen promising targeting ligands based on peptide-protein affinity[18]. Chang and his colleagues established an AI-driven framework for peptide design. This workflow initially employed AlphaFold3 and molecular dynamics (MD) equilibration for iteratively optimized structure generation, subsequently utilizing a transformer-based protein language model for the sequence ranking[15]. Alongside computer-assisted peptide design, they applied SPSS and click chemistry for PML synthesis. SPSS offers several advantages, such as modularity, customizability, and repeatability, which make it favorable for the large-scale production of diverse PMLs[19], and the same is true of click chemistry. Figure 1 shows their collaborative work as a feasible workflow from computational optimization to modular synthesis. It greatly expands the flexibility and diversity of special lipid molecule design.
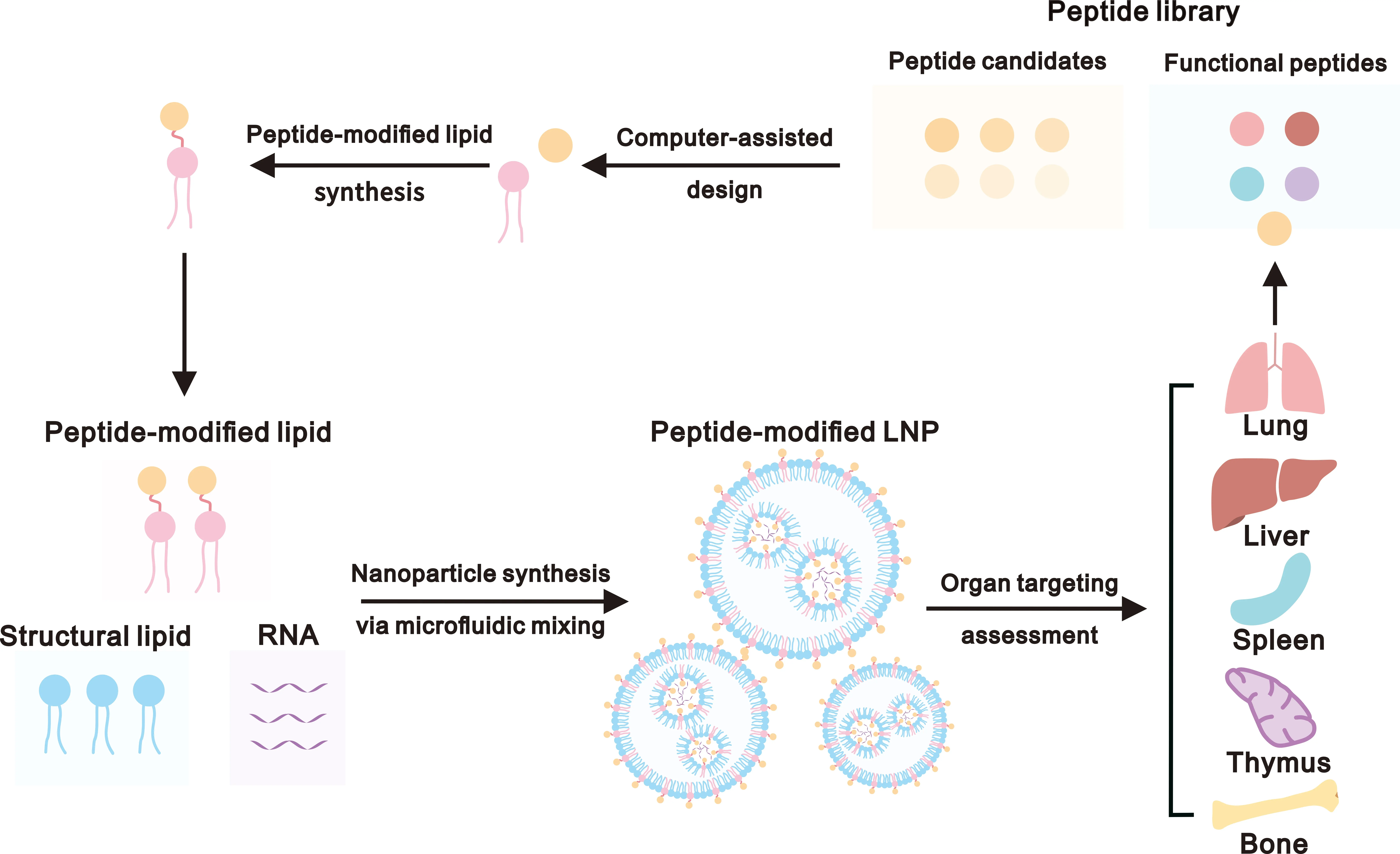
Figure 1. Schematic demonstration of constructing a peptide library for organ-targeted PM-LNPs delivery. This process initiates with various peptide candidates optimized via computer-aided design. The resulting peptides are conjugated to lipids using either SPSS or click chemistry. These PMLs are assembled into nanoparticles with structural lipids and RNA via microfluidic mixing. The resulting PM-LNPs are screened for their delivery efficiency to specific target organs. Successful targeting peptides are incorporated back into the library for subsequent iterative optimization. LNP: lipid nanoparticles; PM-LNPs: peptide-modified LNPs; PMLs: peptide-modified lipids.
3. Factors Affecting the Targeting Properties of PMLs
The targeting performance of PMLs is finely regulated by multiple factors. The composition of the peptide sequence, namely the number, identity, and order of its amino acids, dictates the resultant targeting ability. Lin and his colleagues revealed that subtle changes to single amino acid residues are sufficient to redirect their organ-specific targeting preferences[16] (Figure 2a,b). For instance, modifications with lysines (K) or arginines (R) on the lipid tails direct mRNA to target the lung (> 90% efficacy), whereas the inclusion of glutamate (E), aspartate (D), proline (P), or tryptophan (W) favors spleen delivery (Figure 2c). It is also noteworthy that the integration of alendronate (Ale) achieves 4-fold higher mRNA expression in the bone, and the joint K and Y modification directs LNPs to the thymus with 66.4% specificity[15].
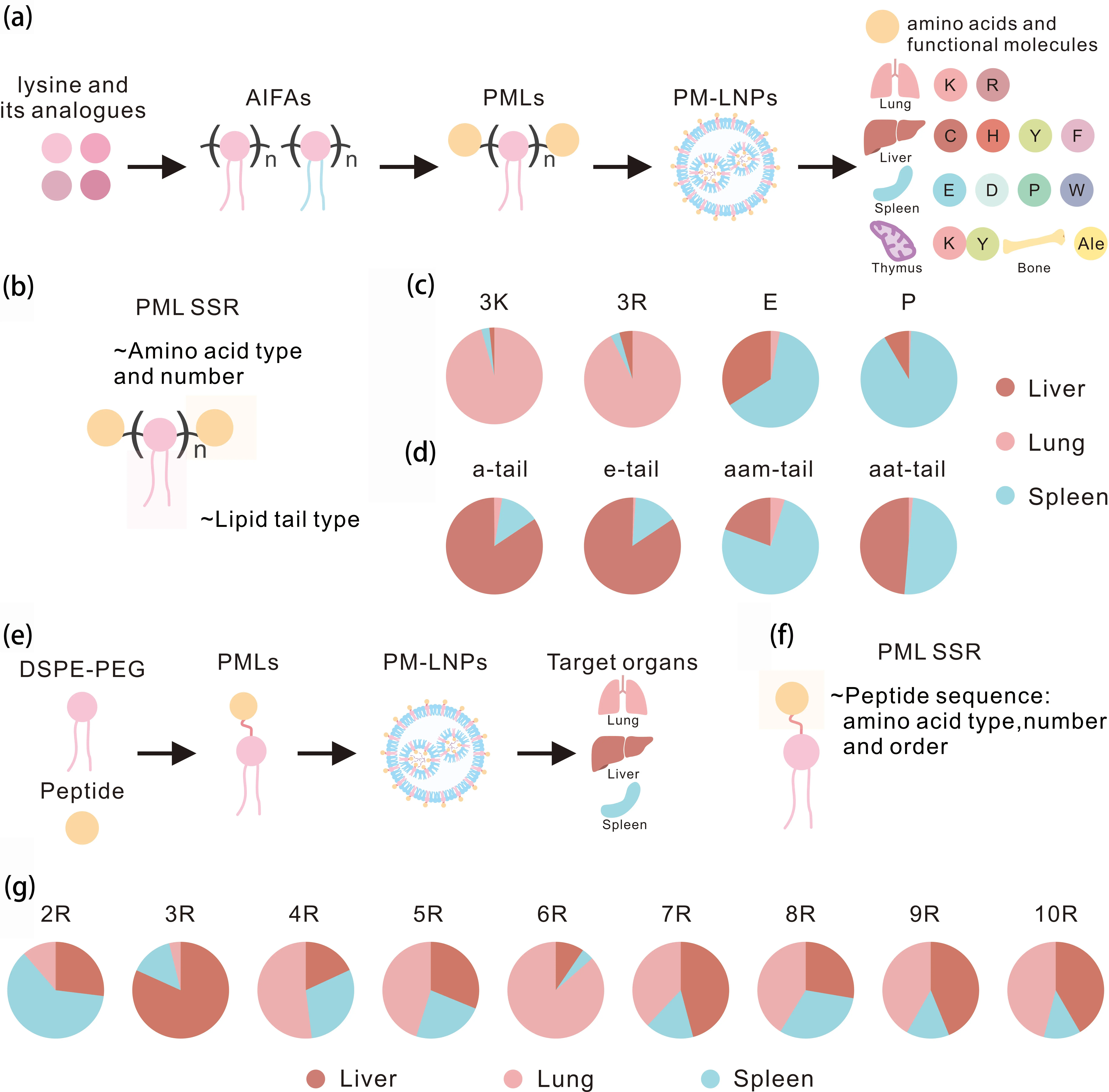
Figure 2. (a) PMLs in Lin’s work, which are composed of AIFAs and diverse amino acids or functional molecules, enable mRNA delivery to specific organs including liver, spleen, lung, thymus and bones; (b) Specific structural characteristics of PMLs in Lin’s work that impact the mRNA delivery efficiency; (c) Relative mRNA expression levels in the liver, spleen, and lung at 6 hours post-treatment of PMLs modified with different amino acids. Amino acids types and numbers are defined as follows: K3, three lysines; R3, three arginines; E, one glutamate; P, one proline; (d) Relative mRNA expression levels in the liver, spleen, and lung after treatment of PMLs with varying tail types. Tail types are defined as follows: a-tails, saturated alkyl chains; e-tails, hydroxylated alkyl chains; aam-tails, amide-containing alkyl chains; aat-tails, ester-containing alkyl chains[16]; (e) PMLs in Chang’s work, which are composed of PEGylated lipids and short peptides, enable mRNA delivery to specific organs; (f) Specific structural characteristics of PMLs in Chang’s work that impact the mRNA delivery efficiency; (g) Relative mRNA expression levels in the liver, spleen, and lung at treatment of PMLs modified with polyarginine peptides ranging from 2 to 10 residues[15]. PMLs: peptide-modified lipids; AIFAs: alkylated Fmoc-protected amino acids; mRNA: messenger RNA; PM-LNPs: peptide-modified LNPs; LNPs: lipid nanoparticles; SSR: structure-sequence relationship; DSPE-PEG: 1,2-distearoyl-sn-glycero-3-phosphoethanolamine–polyethylene glycol.
The targeting specificity of PMLs is not only finely regulated by the peptides but also critically determined by the chemical structure of their lipid tails. Functionally, Figure 2d illustrates that saturated alkyl chains and hydroxyl-containing chains both exhibit a tendency for liver targeting, while amide-containing chains significantly enhance spleen selectivity, achieving up to 86.0% specificity. Ester-containing chains distribute equally between the liver and spleen. Among alkyl chains of different lengths, the 12-carbon chain provides the optimal performance. Consequently, the choice of lipid tails serves as a secondary parameter for tuning the in vivo targeting profile of PMLs[16].
Chang and his colleagues were also investigated the effects of amino acid type on the performance of homopeptides[15]. Beyond that, they identified peptide chain length as another critical determinant: among polyarginine peptides ranging from 2 to 10 residues, 6R exhibits optimal lung-targeting effectiveness, while 3R tends to accumulate preferentially in the liver (Figure 2e,f,g). Meanwhile, they composed spleen-specific hexapeptide sequences using different permutations of D, E and threonine (T). It was found that the DDDDEE sequence yields the highest delivery efficacy, with a 2-fold enhancement compared to other variants[16].
These results collectively demonstrate that the organ selectivity of PMLs exhibits single-amino-acid sensitivity, highlighting the platform’s high tunability. This selectivity is critically dependent on the peptide head, while the lipid tail plays a synergistic role in optimizing targeting efficacy.
3. Different Sights on Mechanism of Organ Selectivity and Efficiency by PMLs
LNP screening typically begins with high-throughput in vitro assays of a designed lipid library. Subsequent in vivo verification allows the identification of lead candidates for clinical trials[20]. However, in vitro performance does not always predict in vivo success, indicating a weak correlation between the two[21]. Lin and his colleagues has conducted research to unveil this discrepancy by analyzing the physical and chemical properties[16]. In vitro, LNPs typically require a relatively high positive charge to enhance cellular internalization and low-to-medium stability for sufficient intracellular mRNA release. In contrast, in vivo delivery demands high stability to ensure the structural integrity of LNPs during systemic circulation and prevent premature cargo leakage. It is also crucial to highlight that, in both in vivo and in vitro environments, high membrane fusion capability is key to achieving efficient endosomal escape, which directly impacts the ultimate mRNA expression[16].
Chang’s work focused on the selective adsorption of plasma proteins onto LNPs[15]. Under in vivo conditions, targeting depends not on positive charge, but on the charge properties conferred by distinct protein corona compositions[22,23]. The protein corona forms based on peptide-protein binding affinity governed by fracture mechanics. After intravenous injection, peptide motifs on LNPs selectively adsorb high-affinity plasma proteins, creating a unique corona that facilitates organ-specific delivery by interacting with cell surface receptors. They verified the above theoretical framework by investigating the connection between peptide 6R, vitronectin, and its cognate receptor, αvβ3 integrin. Starting from the original peptide 6R, the computationally refined peptide RRRYRR shows a 7-fold higher lung signal intensity than that of the template. Thus, the molecular-mechanics-mediated mechanisms act as a robust basis for computer-aided design[15].
4. Conclusions and Future Outlook
Organ-selective mRNA-LNP delivery is crucial for expanding the therapeutic prospects of gene therapies[24], yet it requires a versatile and modular molecular platform to enable efficient extrahepatic targeting. Drawing inspiration from the innate programmability and functional diversity of peptides, Chang’s and Lin’s teams have introduced peptide-modified LNPs as a promising strategy[15,16]. This platform establishes an integrated workflow for the rational design of organ-targeting lipids and provides a solid theoretical basis for improving mRNA-based gene editing therapies in specific tissues. However, several limitations remain for the clinical translation of this strategy. The single-amino-acid sensitivity of the peptide sequence requires higher consistency in mass manufacturing, underscoring the need for a rigorously standardized production process. Furthermore, beyond the in vitro/in vivo disconnect of LNP performance, it is crucial to address the mouse-human discrepancies, especially the differences in protein corona composition. Consequently, utilizing a humanized plasma model is strongly recommended during the design and validation of peptide sequences. Looking ahead, a deeper understanding of these issues will pave the way for a rational research roadmap: first, to develop more clinically viable peptide materials with organ selectivity, and then to expand the scope toward adaptable and programmable functional materials that leverage protein corona-mediated targeting. By leveraging advanced design and synthesis methodologies, we can speed up the building of a comprehensive lipid database. Such a resource would enable the de novo design of targeted delivery systems, fully unleash the potential of AI for large-scale design and screening, and ultimately accelerate the development of LNP-based gene delivery systems.
Authors contribution
Li M: Visualization, writing-original draft, editing.
Li H: Writing, editing, supervision, conceptualization.
Conflicts of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (T2422023).
Copyright
© The Author(s) 2025.
References
-
1. Cullis PR, Felgner PL. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat Rev Drug Discov. 2024;23(9):709-722.[DOI]
-
2. Estapé Senti M, García del Valle L, Schiffelers RM. mRNA delivery systems for cancer immunotherapy: Lipid nanoparticles and beyond. Adv Drug Deliv Rev. 2024;206:115190.[DOI]
-
3. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403-416.[DOI]
-
4. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603-2615.[DOI]
-
5. Song D, Zhao Y, Wang Z, Xu Q. Tuning lipid nanoparticles for RNA delivery to extrahepatic organs. Adv Mater. 2024;36(44):2401445.[DOI]
-
6. Su K, Shi L, Sheng T, Yan X, Lin L, Meng C, et al. Reformulating lipid nanoparticles for organ-targeted mRNA accumulation and translation. Nat Commun. 2024;15(1):5659.[DOI]
-
7. Jeong M, Shin S, Lee G, Lee Y, Park SB, Kang J, et al. Engineered lipid nanoparticles enable therapeutic gene silencing of GTSE1 for the treatment of liver fibrosis. J Control Release. 2024;374:337-348.[DOI]
-
8. Zhang H, Meng C, Yi X, Han J, Wang J, Liu F, et al. Fluorinated lipid nanoparticles for enhancing mRNA delivery efficiency. ACS Nano. 2024;18(11):7825-7836.[DOI]
-
9. Yaghmur A, Østergaard J, Mu H. Lipid nanoparticles for targeted delivery of anticancer therapeutics: Recent advances in development of siRNA and lipoprotein-mimicking nanocarriers. Adv Drug Deliv Rev. 2023;203:115136.[DOI]
-
10. Geisler HC, Ghalsasi AA, Safford HC, Swingle KL, Thatte AS, Mukalel AJ, et al. EGFR-targeted ionizable lipid nanoparticles enhance in vivo mRNA delivery to the placenta. J Control Release. 2024;371:455-469.[DOI]
-
11. Levin A, Hakala TA, Schnaider L, Bernardes GJL, Gazit E, Knowles TPJ. Biomimetic peptide self-assembly for functional materials. Nat Rev Chem. 2020;4(11):615-634.[DOI]
-
12. Zhang Z, Baxter AE, Ren D, Qin K, Chen Z, Collins SM, et al. Efficient engineering of human and mouse primary cells using peptide-assisted genome editing. Nat Biotechnol. 2023;42(2):305-315.[DOI]
-
13. Ledford BT, Akerman AW, Sun K, Gillis DC, Weiss JM, Vang J, et al. Peptide amphiphile supramolecular nanofibers designed to target abdominal aortic aneurysms. ACS Nano. 2022;16(5):7309-7322.[DOI]
-
14. Wang MD, Lv GT, An HW, Zhang NY, Wang H. In situ self‐assembly of bispecific peptide for cancer immunotherapy. Angew Chem Int Ed Engl. 2022;61(10):e202113649.[DOI]
-
15. Chang T, Zheng Y, Jiang M, Jia S, Bai J, Zheng Z, et al. Peptide codes for organ-selective mRNA delivery. Nat Mater. 2025.[DOI]
-
16. Lin Y, Li M, Luo Z, Meng Y, Zong Y, Ren H, et al. Tissue-specific mRNA delivery and prime editing with peptide–ionizable lipid nanoparticles. Nat Mater. 2025.[DOI]
-
17. Koh HY, Zheng Y, Yang M, Arora R, Webb GI, Pan S, et al. AI-driven protein design. Nat Rev Bioeng. 2025.[DOI]
-
18. Huang J, Xu Y, Xue Y, Huang Y, Li X, Chen X, et al. Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences. Nat Biomed Eng. 2023;7(6):797-810.[DOI]
-
19. Lin P, Xu Y, Bali SK, Kim J, Gimeno A, Roberts ET, et al. Solid‐phase‐supported chemoenzymatic synthesis and analysis of chondroitin sulfate proteoglycan glycopeptides. Angew Chem Int Ed. 2024;63(34):e202405671.[DOI]
-
20. Herrera-Barrera M, Ryals RC, Gautam M, Jozic A, Landry M, Korzun T, et al. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci Adv. 2023;9(2):eadd4623.[DOI]
-
21. Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, et al. A direct comparison of in vitro and in vivo nucleic acid delivery mediated by hundreds of nanoparticles reveals a weak correlation. Nano Lett. 2018;18(3):2148-2157.[DOI]
-
22. Dilliard SA, Cheng Q, Siegwart DJ. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc Natl Acad Sci U.S.A. 2021;118(52):e2109256118.[DOI]
-
23. Miao L, Lin J, Huang Y, Li L, Delcassian D, Ge Y, et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat Commun. 2020;11(1):2424.[DOI]
-
24. Parhiz H, Atochina-Vasserman EN, Weissman D. mRNA-based therapeutics: looking beyond COVID-19 vaccines. Lancet. 2024;403(10432):1192-1204.[DOI]
Copyright
© The Author(s) 2025. This is an Open Access article licensed under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, sharing, adaptation, distribution and reproduction in any medium or format, for any purpose, even commercially, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Publisher’s Note
Share And Cite



